Tools for Neuroscience
Fall 2015 - Spring 2024
Description
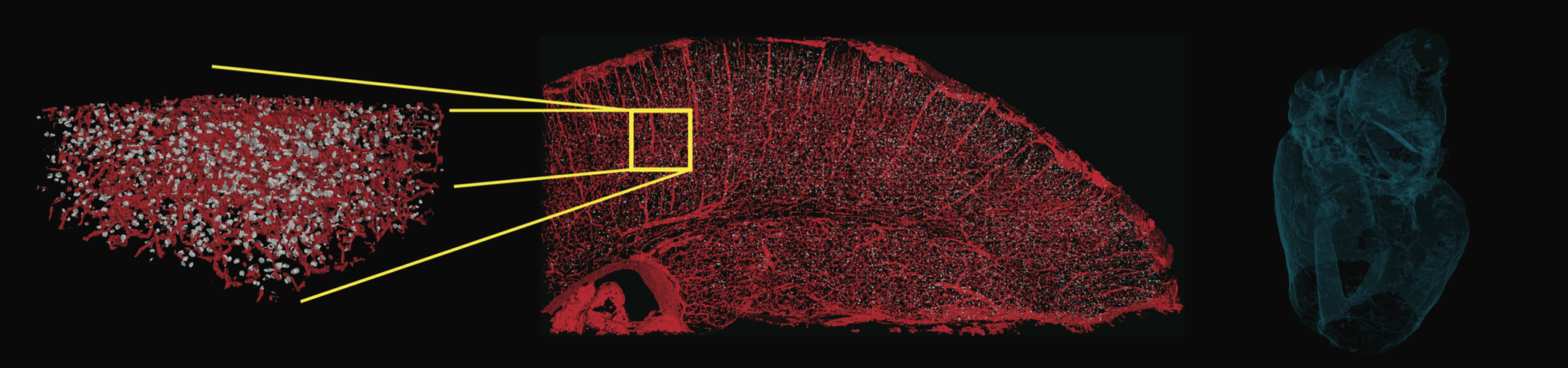
We, and our partners spread across multiple institutions around the country, are studying the function of the brain, directly observing it at work, and tracking its activity in real-time as an animal learns. Combining state-of-the-art automated training procedures with next generation temporal focusing two-photon calcium imaging techniques, we are measuring how the activity of thousands of neurons in the brain correspond to an animal's experience of the visual world. By watching these patterns evolve over the course of learning, we can capture powerful clues about the learning rules that the brain uses to grasp new information. To dig deeper into the machinery of the brain, we are subsequently deploying massive-scale imaging techniques like synchrotron X-ray tomography on the same brains to image and reconstruct the connectome of a significant portion of the rodent's visual cortex. To date, a combined effort to characterize the function and anatomy of such a large portion of a brain has never been attempted.
Collecting data from brains is one thing, but how do we translate that data into a useful algorithm to solve a real problem? A crucial question for a neural network model is: does it have the same structure and function as networks of cells in the brain? The answer to this seminal question is extremely complicated. But with nature as our guide, we have a solid reference point in the connectome.
An understanding of the connectome is fundamental, long-term research. The remarkable diversity of life on planet earth means that there are many aspects of sensory systems and associated neural computations to explore. This is exactly why computer scientists are interested in this research – it provides new ways to think about problem solving and models of computation. While the clinical goals of this research are admirable, the promise of a new model of computation could be even more important. While current artificial intelligence software is impressive, it still comes up short in the contexts that matter most to the healthcare, defense, and intelligence communities: settings where decisions must be made from limited amounts of ambiguous data, and where those decisions must account for uncertainty. It is in precisely these contexts that humans excel. The human brain, arguably the most powerful known computational system, evolved to operate with extraordinary efficiency and accuracy in ambiguous, complicated environments. Initial work in building biologically-inspired neural networks has shown great promise, but the structure of today's artificial neural networks is a pale reflection of the complexity found in even relatively simple mammalian brains.
This work was supported by IARPA contract #D16PC00002, the Department of Defense (Army Research Laboratory) under the contract W911NF-18-1-0292, and the NVIDIA Corporation
Publications
- "Perceptography Unveils the Causal Contribution of Inferior Temporal Cortex,,,,,
to Visual Perception,"Nature Communications,April 2024.[pdf][bibtex]@article{shahbazi2024perceptography,
title={Perceptography Unveils the Causal Contribution of Inferior Temporal Cortex
to Visual Perception},
author={Shahbazi, Elia and
Ma, Timothy and
Pernu{\v{s}}, Martin and
Scheirer, Walter and
Afraz, Arash},
journal={Nature Communications},
volume={15},
number={1},
pages={3347},
year={2024},
publisher={Nature Publishing Group UK London}
}
- "An Assistive Computer Vision Tool to Automatically Detect Changes in Fish,, , , , ,
Behavior In Response to Ambient Odor,"Scientific Reports,January 2021.[pdf] [code][bibtex]@article {Banerjee_SciRep2021,
author = {Banerjee, Sreya and Alvey, Lauren and Brown, Paula and Yue,
Sophie and Li, Lei and Scheirer, Walter J.},
title = {An Assistive Computer Vision Tool to Automatically Detect
Changes in Fish Behavior In Response to Ambient Odor},
year = {2021},
volume = {11},
number = {21},
journal = {Scientific Reports}
}
- "Flexible Learning-Free Segmentation and Reconstruction for Neuronal Circuit Tracing,", , , , , ,
, , , , ,
,Scientific Reports,September 2018.[pdf] [code][bibtex]@article {Shahbazi278515,
author = {Shahbazi, Ali and
Kinnison, Jeffery and
Vescovi, Rafael and
Du, Ming and
Hill, Robert and
J{\"o}sch, Maximilian and
Takeno, Marc and
Zeng, Hongkui and
da Costa, Nuno Ma{\c c}arico and
Grutzendler, Jaime and
Kasthuri, Narayanan and
Scheirer, Walter J.},
title = {Flexible Learning-Free Segmentation and Reconstruction for Neuronal Circuit Tracing},
journal = {Scientific Reports}
volume = {8},
number = {14247},
year = {2018},
}
- "Neuron Segmentation Using Deep Complete Bipartite Networks,", , , ,Proceedings of the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI),September 2017.[pdf][bibtex]@InProceedings{Chen_2017_MICCAI,
author = {Jianxu Chen and Sreya Banerjee and Abhinav Grama and Walter J. Scheirer and Danny Z. Chen},
title = {Neuron Segmentation Using Deep Complete Bipartite Networks},
booktitle = {International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI)},
month = {September},
year = {2017}
}
- "Reconstruction of Genetically Identified Neurons Imaged by Serial-Section Electron Microscopy,", , , , , ,, , , ,eLife,July 2016.[pdf][bibtex]@article{Joesch_2016_eLife,
author = {Joesch, Maximilian and Mankus, David and Yamagata, Masahito and Shahbazi, Ali and Schalek, Richard and
Suissa-Peleg, Adi and Meister, Markus and Lichtman, Jeff W. and Scheirer, Walter J. and Sanes, Joshua R.},
title = {Reconstruction of Genetically Identified Neurons Imaged by Serial-Section Electron Microscopy},
journal = {eLife},
volume = {5},
pages = {e15015},
year = {2016}
}